Sternberg PW, et al. (2024). WormBase 2024: status and transitioning to Alliance infrastructure. Genetics, Apr 4
Wright A, et al. (2024). FAIR Header Reference genome: a TRUSTworthy standard. Brief Bioinform, Mar 27
Alliance of Genome Resources Consortium. (2024). Updates to the Alliance of Genome Resources Central Infrastructure. Genetics, Mar 29
De Jesus Martinez, T et al. (2023). JBrowse Jupyter: a Python interface to JBrowse 2. Bioinformatics, Jan 1;39(1)
Diesh C, et al. (2023). JBrowse 2: a modular genome browser with views of synteny and structural variation. Genome Biol. Apr 17;24(1):74
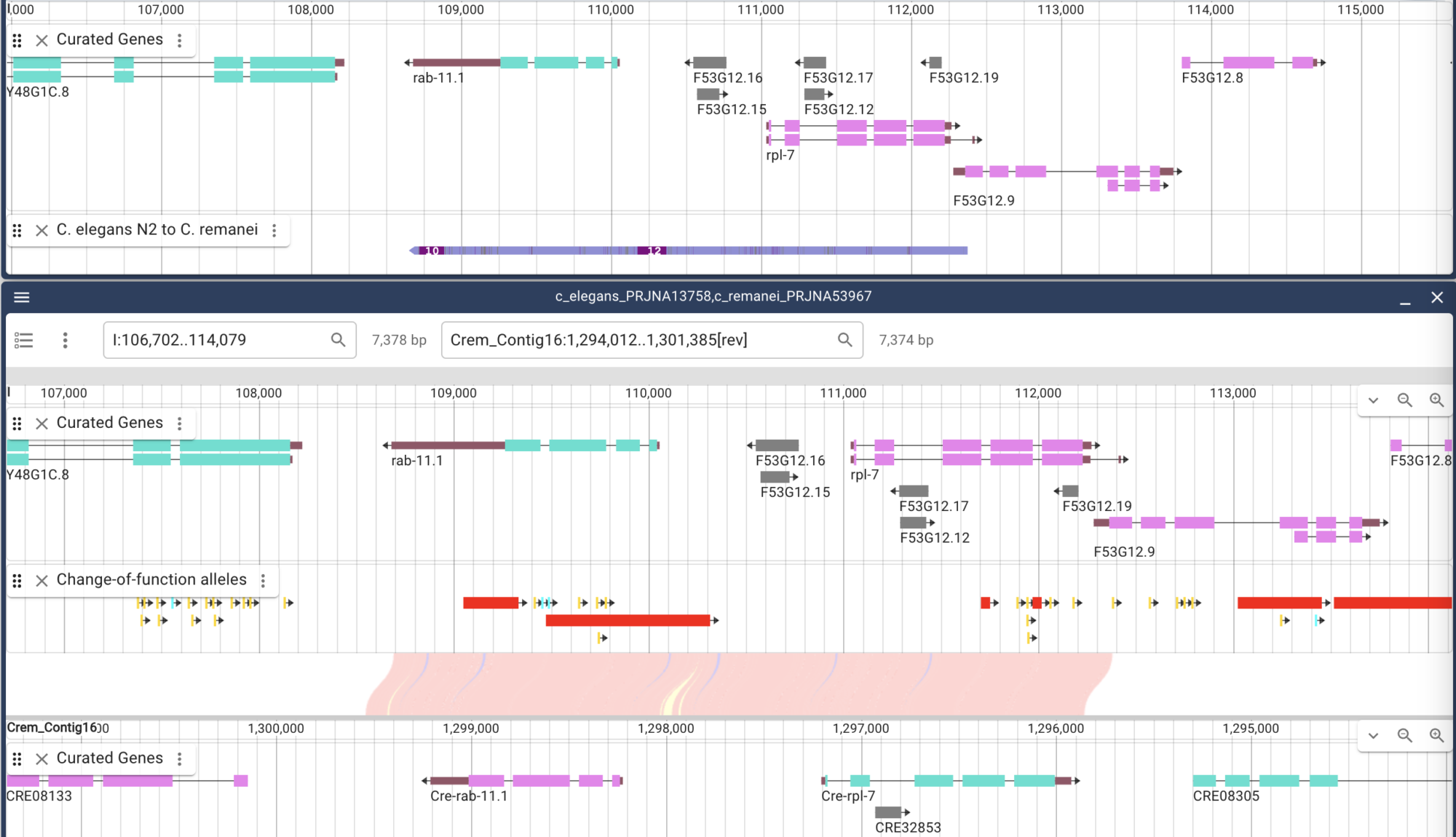
Davis, P, et al. (2022) WormBase in 2022-data, processes, and tools for analyzing Caenorhabditis elegans. Genetics, Apr 4;220(4)
Harris, TW et al. (2020). WormBase: a modern Model Organism Information Resource. Nucleic acids research, 48
Alliance of Genome Resources Consortium (2020). Alliance of Genome Resources Portal: unified model organism research platform. Nucleic acids research, 48
Lee, R et al. (2018). WormBase 2017: molting into a new stage. Nucleic acids research, 46
Grove, C et al. (2018). Using WormBase: A Genome Biology Resource for Caenorhabditis elegans and Related Nematodes. Methods in molecular biology (Clifton, N.J.), 1757, 399–470.
Howe KL et al. (2016) WormBase 2016: expanding to enable helminth genomic research. Nucleic Acids Research 2016 Jan 4
Jung S et al. (2011) The Chado Natural Diversity module: a new generic database schema for large-scale phenotyping and genotyping data. Database (Oxford) 2011 Nov 26
McKay S and Cain S (2009) Genome Browsers. In Bioinformatics: Tools and Applications (pp. 39-63) New York, NY: Springer.
O’Connor BD et al. (2008) GMODWeb: a web framework for the Generic Model Organism Database. Genome Biol 9: R102
Arnaiz O et al. (2007) ParameciumDB: a community resource that integrates the Paramecium tetraurelia genome sequence with genetic data. Nucleic Acids Res 35: D439-44
Harrington JJ et al. (2001) Creation of genome-wide protein expression libraries using random activation of gene expression. Nat Biotechnol 19: 440-5
Cain SJ & Chau PC (1998) Transition probability cell cycle model with product formation. Biotechnol Bioeng 58: 387-99
Cain SJ & Chau PC (1997) Transition probability cell cycle model. Part I–Balanced growth. J Theor Biol 185: 55-67
Cain SJ & Chau PC (1997) Transition probability cell cycle model. Part II–Non-balanced growth. J Theor Biol 185: 69-79
Cain SJ & Chau PC (1997) A transition probability cell cycle model simulation of bivariate DNA/bromodeoxyuridine distributions. Cytometry 27: 239-49