Education
Doctor of Philosophy in Chemical Engineering, 1997
University of California at San Diego
Dissertation title: A Transition Probability Model Analysis of Cell Cycle Dependent Protein Production
Bachelor of Science in Chemical Engineering, Summa Cum Laude, 1990 The Ohio State University
Languages and Platforms
Perl, Python, React and Dojo (JavaScript), Java, PHP, Fortran, C++
Docker, PostgreSQL, MySQL, Galaxy, InterMine, Tripal, Drupal, WordPress, MediaWiki
Work Experience
Sole proprietor (primarily subcontracting for the Ontario Institute for Cancer Research) 10/2008 to present
Current projects include:
● GMOD Project Manager 10/2008 to present
GMOD responsibilities include:
- Directing software development and coordinating the efforts of many paid and volunteer programmers at several academic, non-profit and corporate institutions in a variety of application modules that are part of the Generic Model Organism Database project. Coordination takes place in a variety of venues, including GMOD meetings, web site communications, teleconferences, email, and conference workshops.
- Lead software developer of the Chado project: the central database schema of GMOD; it is a highly normalized, ontology-dependent schema that encompasses several domains in biology. Contributing developer for JBrowse, GBrowse, Apollo, and BioPerl projects.
- Developing instructional materials for and teaching a variety of GMOD courses, including “GMOD Summer Schools” (an intensive course covering several GMOD applications) “GMOD in the Cloud” (using Amazon Web Services with GMOD appliances), and many JBrowse workshops, including JBrowse-Jupyter Python notebooks.
- Additional contract work supporting organizations with Chado/GMOD-related activities.
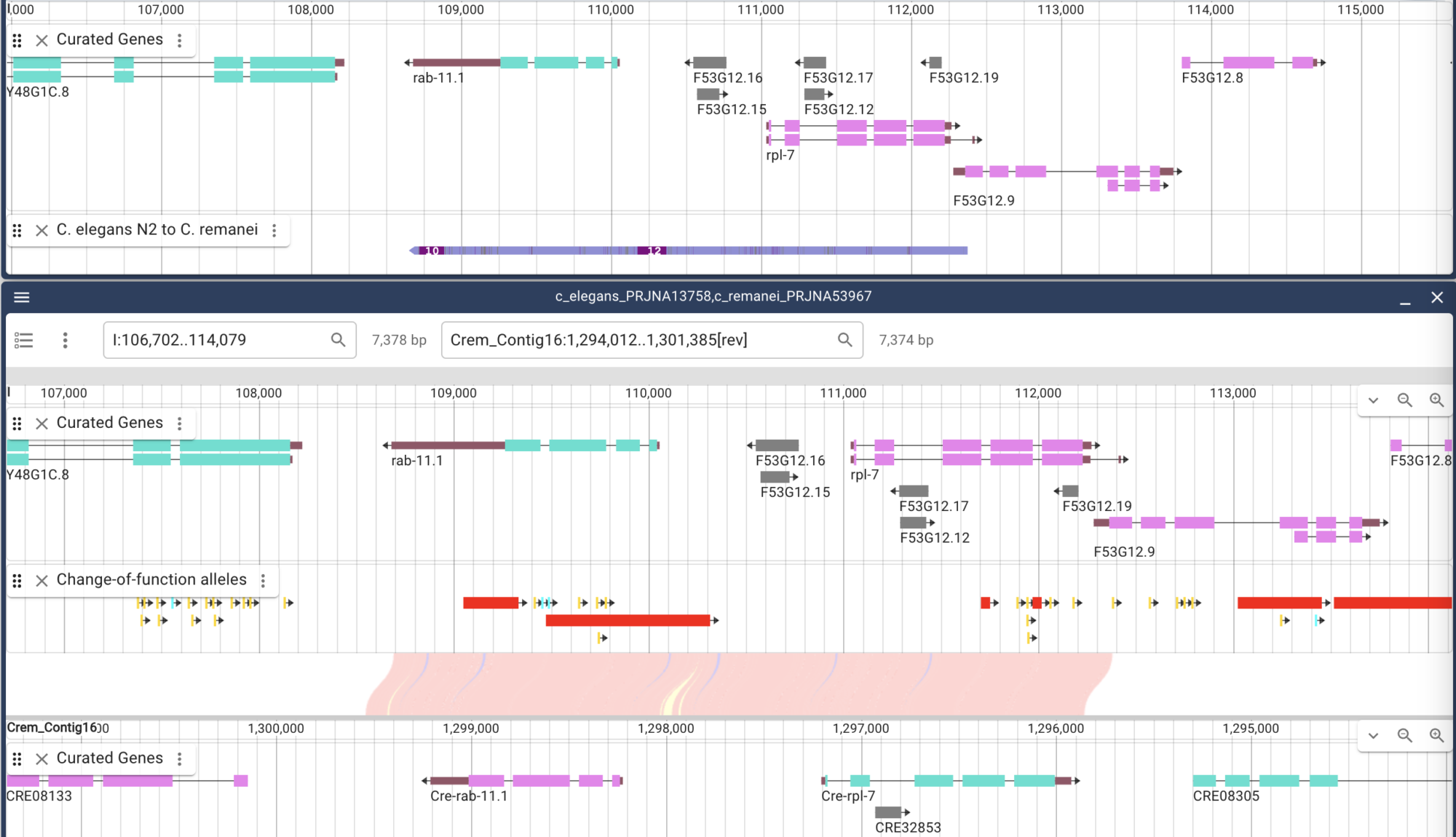
● Senior WormBase Developer 8/2014 to present
Primarily responsible for genome browsing tools, both maintaining existing and developing new tools and pipelines involving Docker, Javascript, shell scripts and Perl at WormBase. Support website development in Template Toolkit, Moose and Perl. Support development in InterMine.
● Alliance for Genome Resources Working Group Leader 6/2017 to present
Leading the group of developers responsible for all aspects of the Alliance website that relate to ingest and display of genome feature data including genes and variants across all participating species. This involved developing internal GFF3 standards and custom JavaScript widgets for displaying data as well as data analysis pipelines and JBrowse instances that are consistent across participating species. Participate in scrum/agile groups as product owner and specialist.
● VirusSeq Product Manager 9/2021 to present
Manage developer resources and communication with users for VirusSeq, the SARS-CoV-2 sequence repository for Canada.
● Human Cancer Models Initiative Product Manager 4/2022 to present
Manage developer resources and communication for HCMI, a searchable catalog of cancer model systems.
Faculty Member 11/2002 to 8/2016
University of Phoenix, Cleveland Campus, Independence, OH
Taught several ground, Flexnet (blended), and online classes in the BSIT program, including courses in database design and applications, web development, and programming.
GMOD Project Coordinator 9/2002 to 9/2008
Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
Same responsibilities as “GMOD Project Manager” as above.
Project Leader/Lead Bioinformatics Developer 6/1999 to 5/2002
Athersys, Inc., Cleveland, OH
Responsibilities included supervising the work of two scientific software developers; collecting business requirements from involved scientists and creating functional specifications for internal application development; creating and maintaining scientific software and databases; solving all data analysis problems. Major accomplishments of the group included:
- Designing, implementing and maintaining a company-wide intranet using MS SQL Server, Java applets and Visual Basic script/ASP.
- Designing, implementing and maintaining a data analysis pipeline and relational data repository using MS SQL Server, Perl, Visual Basic, Java, JavaScript and a network of Unix and NT workstations; front-end display via Active Server Pages (ASP).
- Designing, implementing and maintaining a laboratory information management system (LIMS) for a DNA sequencing lab using MS SQL Server, Visual Basic, Visual C++, and Perl; front-end display via ASP and Java applets.
- Designing, implementing and maintaining a mixed computing cluster (PC/Linux, Alpha/Linux, PC/Solaris, Sun/Solaris) running Platform Computing’s LSF.
- Designing, implementing and maintaining a distributed computing network to allow desktop PCs to contribute to the computing cluster.
- Identifying thousands of novel genes and tentatively predicting their function as part of a large scale gene discovery effort.
Pre- and Post-Sales Engineer 2/1998 to 5/1999
Pangea Systems, Inc (later known as DoubleTwist Inc), Oakland, CA
Responsibilities included providing sales and technical support in both the science and software of bioinformatics; demonstrating software to prospective customers, including LIMS, pipeline, and pathways related products; guiding customers in determining the hardware and software requirements to fit their needs; performing on site installation of databases and software; performing bug fixes on Java-based source code.
Customer Support Programming Specialist/Life Sciences Development 1/1997 to 2/1998
Molecular Simulations, Inc (later known as Accelrys, Inc), San Diego, CA
Responsibilities included creating modules to customer specifications for molecular modeling software (Cerius2) using its software developers kit, creating browser interfaces to server-side Cerius2, translating existing modules between FORTRAN and C/C++, demonstrating the use of the modules to customers, and developing tutorials, example applets and documentation for a Java Bean-based software developers kit for a browser-based bioinformatics application.
Teaching Assistant 9/1990 to 5/1997
University of California at San Diego, La Jolla, CA
Classes included: BASIC Programming for non-science majors, Chemistry for non-science majors, Introductory Chemistry, Introductory Chemistry Lab, Analytical Chemistry Lab, Process Control, Heat Transfer, Unit Operations, Unit Operations Lab.